Biomarker ID and Analysis
Direct Analysis
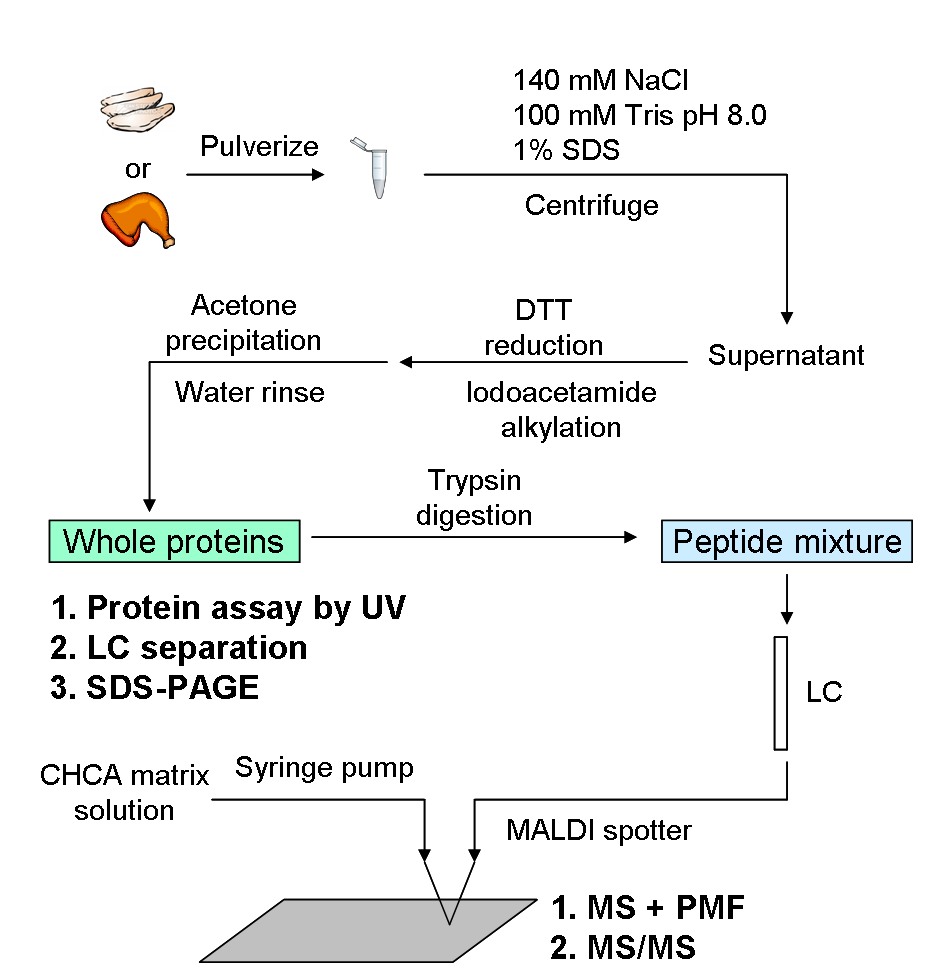
In the simplest case, complex samples are spotted directly onto a MALDI plate. If the sample consists of proteins that are digested with trypsin, then the most abundant proteins can be identified by Peptide Mass Fingerprinting (PMF) starting from high-resolution reflectron spectra. SimulTOF Systems has developed special PMF software that takes advantage of the well-known preferential detection of arginine-containing peptides that makes PMF practical even for whole tissue preparations, so long as most proteins are largely intact. Differences between preparations can be determined using peak list information loaded into spreadsheets like Excel, and/or exported to 3rd party software like R, SpotFire or GeneSpring. The following poster describes this workflow starting with chicken meat preparations obtained from a grocery store, which is a model system for studying myosin heavy chain differences in human muscle biopsy samples.
Chicken White Meat vs Dark Meat by Peptide Mass Fingerprinting—ASMS 2011

LC MALDI
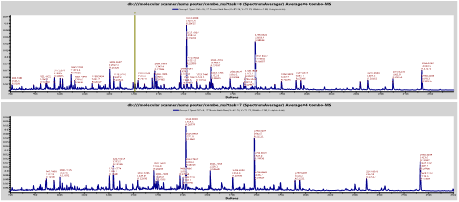
In a second workflow, the same tryptic digests are separated by reversed phase HPLC and directly deposited along with matrix onto MALDI plates. Even one round of peptide separation trypically increases the number of detectable components by 2 orders of magnitude. When internal standards are added to each spot, most precursor masses are measured to within a few ppm. The resulting peak lists, containing thousands of masses, can be directly used for peptide mass fingerprinting, resulting in the identification of > 30 proteins from yeast preparations. These protein assignments and hundreds more are typically assigned following TOF-TOF analysis. The following poster shows the evidence for the correct identification of abundant protein identification by direct PMF analysis of chicken muscle tryptic digests and describes what differences were found between chicken white meat and dark meat.
Chicken White Meat vs Dark Meat by LC-MALDI with MS-MS—ASMS 2011
Immunoaffinity
In the third approach, the proteins of interest are first captured by affinity chromatography (Mass Spectrometric Immunoassay (see Mass Spectrometric Immunoassay revisited), followed by direct MALDI analysis of intact proteins. The same ‘pulldown’ technology can also be applied to determine protein interaction partners. Immunoaffinity techniques can also be applied to determine quantitative changes in protein expression based on immunoaffinity capture of specific peptides (see Precision of Heavy−Light Peptide Ratios Measured by MALDI-TOF).
Atomic Composition Analysis
Reflectron MALDI mass spectra can easily be collected at very high resolution. With internal standards, this often results in sufficient precision to determine an exact chemical composition. MALDI MS-MS spectra acquired using the SimulTOF 300 MALDI-TOF-TOF can also be compared directly to existing MS-MS spectrum libraries, for example, those at the National Institute for Standards and Technology.

